fMRI Brain Network Construction
The fmri brain network construction is a BIDS-formatted fMRI data processing pipeline for general researchers to build their own functional brain network from fMRI data. We use the existing ABCD-HCP BIDS fMRI Pipeline, which combines complicated FSL, Freesurfer, and Linux commands into one docker command. The detailed pipeline description could be found at ABCD-BIDS Community Collection (ABCC).
Preparation:
System requirement:
- Linux or MacOS (Intel Core)
Requirements
- Docker or Singularity
- All tools used would be downloaded and configured inside of
DockerandSingularity
- All tools used would be downloaded and configured inside of
- FreeSurfer license
- DICOM to NIFTI tools (eg. MRIcroGL)
Install the Pipeline
- With Docker
docker pull dcanumn/abcd-hcp-pipeline - With Singularity
singularity pull docker://dcanumn/abcd-hcp-pipeline singularity build abcd-hcp-pipeline.sif docker://dcanumn/abcd-hcp-pipeline
Run the Pipeline
-
The raw data should be in BIDS format. Take the ABCD dataset as an example using the subject (NDARINV003RTV85)
- Make sure you have access to ABCD data on NDA
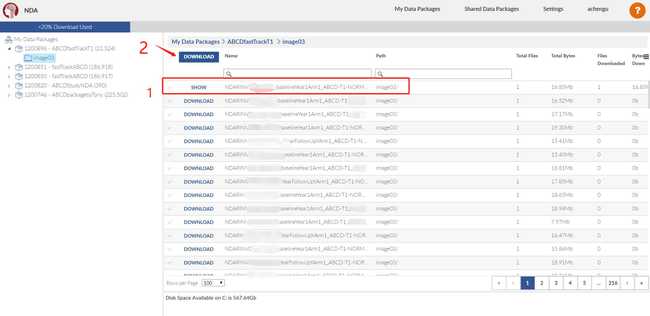
- Create your own data package by following the numbers.
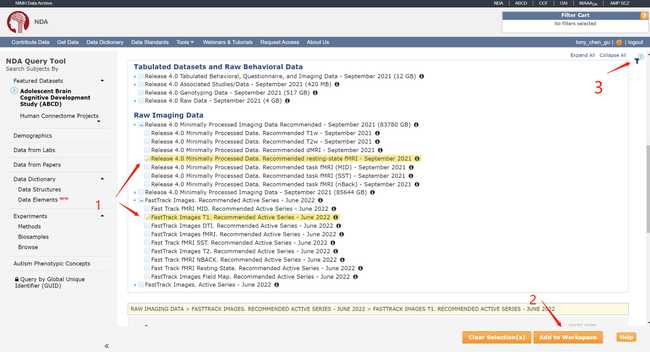
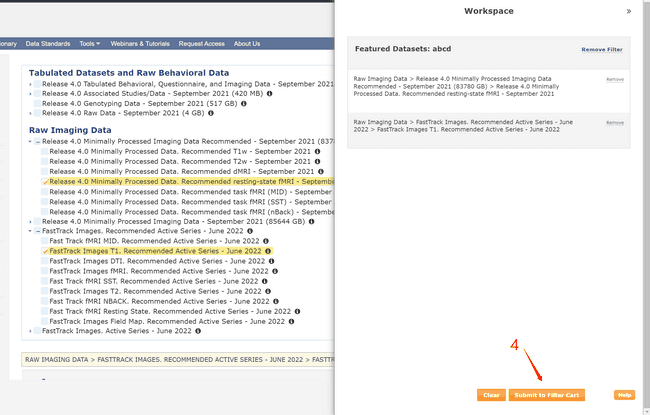
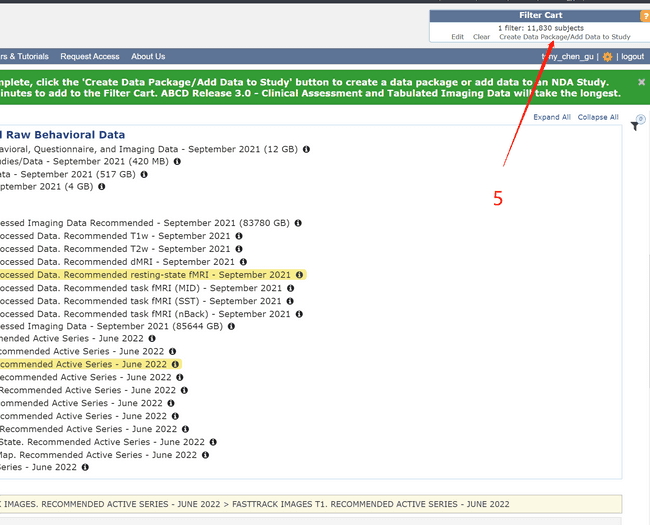
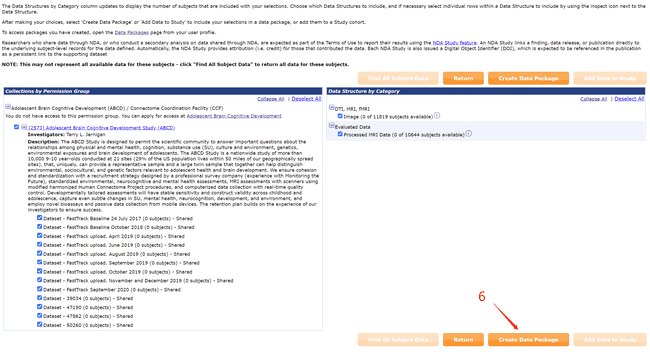
- Using the NDADownLoadManager to download the raw T1 images (ABCD-T1-NORM_run-20181001100823) and processed fmri and T1 images.
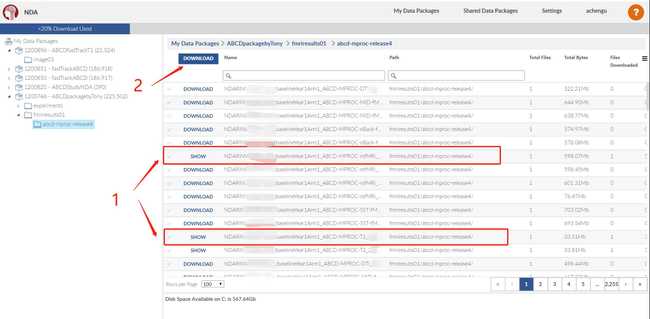
- The processed fmri and T1 images are already in BIDS format, so we could directly combine two folders.
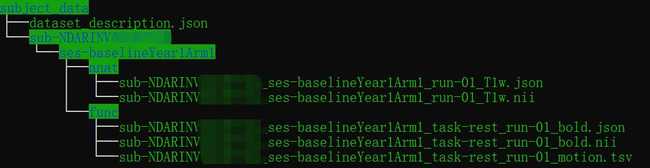
- Extract the raw T1 image and convert it to .nii. Using any dcm to nii tools (eg. MRIcroGL) to convert the raw data (ABCD-T1-NORM_run-20181001100823) to nii.
- We need its json file to supplement processed T1 json. Using the converted json to supplement the processed T1 json according to the following image.
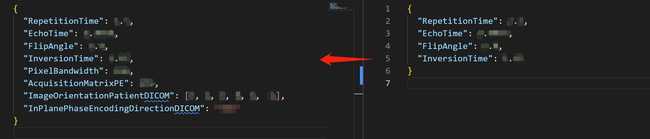
-
With Docker
docker run --rm \ -v /path/to/bids_dataset:/bids_input:ro \ -v /path/to/outputs:/output \ -v /path/to/freesurfer/license:/license \ dcanumn/abcd-hcp-pipeline /bids_input /output --freesurfer-license=/license.txt [OPTIONS] -
With Singularity
env -i singularity run \ -B /path/to/bids_dataset:/bids_input \ -B /path/to/outputs:/output \ -B /path/to/freesurfer/license.txt:/opt/freesurfer/license.txt \ ./abcd-hcp-pipeline.sif /bids_input /output --freesurfer-license=/opt/freesurfer/license.txt [OPTIONS] -
My Docker command:
docker run --rm \ -v /mnt/d/ABIDE/full_steps/subject_data_old:/bids_input:ro \ -v /mnt/d/ABIDE/full_steps/latest_output:/output \ -v /home/chentony2011/license.txt:/license \ dcanumn/abcd-hcp-pipeline /bids_input /output --freesurfer-license=/license --ncpus 15 -
If you met error message like:
Exception: error caught during stage: DCANBOLDProcessingtry version v0.1.0 of the pipeline. With command
-stage DCANBOLDProcessingto specify the starting stage.docker run --rm \ -v /mnt/d/ABIDE/full_steps/subject_data_old:/bids_input:ro \ -v /mnt/d/ABIDE/full_steps/latest_output:/output \ -v /home/chentony2011/license.txt:/license \ dcanumn/abcd-hcp-pipeline:v0.1.0 /bids_input /output --freesurfer-license=/license --ncpus 15 --stage DCANBOLDProcessing
Generate Network
- The pipeline above won't generate the network automatically. You will need to run the script
generate_network.pyto generate the network. - The pipeline output is in the structure:
output_dir/ |__ sub-id |__ ses-session |__ files | |__DCANBOLDProc<ver> | |__summary_DCANBOLDProc<ver> | | |__executivesummary | |__ MNINonLinear | | |__ fsaverage_LR32k | | |__ Results | |__ T1w | | |__ id | |__[T2w] | |__ task-<taskname> |__ logs - The file used to generate brain network is placed in the folder
output_dir/sub-id/ses-session/files/MNINonLinear/Results/. All files ends with.ptseries.niican be used to generate the network. - To generate network put the script provided
generate_network.pyin the folder and run it. The output network would be saved as a.matfile in a folder namedconnectivity_output. - You could change the input folder, output folder, and the connectivity measure in the code.
More Options
```shell
Positional Arguments:
bids_dir Path to the BIDS data.
output_dir Path to the output directory.
Optional Arguments:
-h / --help Show the function help and exit.
-v / --version Show the version of the package and exit.
--participant-label ID [ID...]
Specify the participant ID desired to run.
Default is to run all participant in the directory.
--session-id SESSION_ID [SESSION_ID ...]
Specify the Session ID desired to run. (Exclude "ses-")
Default is to run all participant in the directory.
--freesurfer-license LICENSE_FILE
--all-sessions Merge all sessions into one for each subject.
--ncpus NCPUS
Number of cores to run the pipeline.
--stage STAGE
Specified the desired substage to run.
--bandstop LOWER UPPER
Motion regressor band-stop filtering parameter.
--custom-clean JSON
Run the DCAN automatic cleaning script based on the
specified file structure provided after the pipeline.
--study-template HEAD BRAIN
Head and brain template for intermediate
nonlinear registration and masking.
--ignore {func,dwi} Ignore the modality in processing.
Runtime Arguments: (For debugging use)
--check-outputs-only Check the existence of the output.
--print-commands-only Only print out the command used to run each stage.
--ignore-expected-outputs
Continue the pipeline with missing expected output.
```